Overview
Aligning data from different domains is fundamental in machine learning, from single-cell multiomics to neural latent space alignment. This work introduces a learnable Gromov-Wasserstein framework that transforms the computationally intractable quadratic assignment problem into a scalable, inductive solution.
Rather than solving Gromov-Wasserstein (GW) problems directly through iterative optimization, we learn embeddings that map both domains into a common space where a single optimal transport problem produces the alignment. This amortized approach enables:
- Inductive generalization: Train on small samples, apply to arbitrarily large test sets
- Superior scalability: Handle 45,000+ samples where standard solvers fail beyond 25,000
- Flexible extensions: Support non-metric structures through rank-based matching
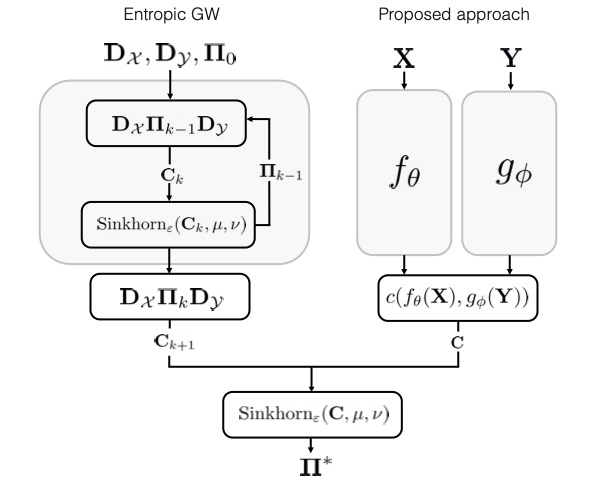
Method: Learning Alignment Through Bilevel Optimization
From Quadratic to Linear Assignment
The Gromov-Wasserstein problem seeks to align metric spaces by minimizing distance disagreement:
\[d^2_{\text{GW}} = \min_{\mathbf{\Pi} \in U(\mu, \nu)} \| \mathbf{D}_\mathcal{X} - \mathbf{\Pi} \mathbf{D}_\mathcal{Y} \mathbf{\Pi}^\T \|_\mathrm{F}^2\]This is a quadratic assignment problem (NP-hard). Instead of solving it directly, we pose a bilevel optimization:
\[\begin{aligned} \Vec{\Pi}^* =& \argmin_{\theta, \phi} \, \left\| \mathbf{D}_\mathcal{X} - \Vec{\Pi}(\theta, \phi) \mathbf{D}_\mathcal{Y} \Vec{\Pi}^\T(\theta, \phi) \right\|_{\mathrm{F}}^2 \\ & \,\, \text{s.t.} \,\, \Vec{\Pi}(\theta, \phi) = \argmin_{\Vec{\Pi} \in U(\mu, \nu)} \langle \mathbf{\Pi}, {c}(f_\theta(\mathbf{X}), g_\phi(\mathbf{Y}))\rangle \end{aligned}\]Here, $f_\theta$ and $g_\phi$ are neural networks that embed samples from $\mathcal{X}$ and $\mathcal{Y}$ into a common space. The inner problem is a linear optimal transport problem, efficiently solvable via Sinkhorn algorithm.
Key Insight: Amortization
By learning the embeddings, we amortize the iterative GW computation. At inference:
- Embed new samples through learned $f_\theta$ and $g_\phi$
- Solve a single entropy-regularized OT problem
- No need to recompute geodesic distances or iterate through GW loops
This is implemented using implicit differentiation to backpropagate gradients through the OT solver, enabling end-to-end learning.
Extensions: Making It Practical
1. Rank-Based Matching for Non-Metric Structures
Standard GW requires metric spaces. For general dissimilarities (e.g., different biological modalities with incompatible scales), we match ranks instead of distances:
\[\min_{\theta, \phi} \left\| \mathcal{R}_\delta\left(\mathbf{D}_\mathcal{X}\right) - \mathcal{R}_\delta\left(\mathbf{\Pi}(\theta, \phi) \mathbf{D}_\mathcal{Y} \mathbf{\Pi}^\T(\theta, \phi) \right) \right\|_{\mathrm{F}}^2\]where $\mathcal{R}_\delta$ is a differentiable soft-ranking operator. This makes the method:
- Scale-invariant: Handles point clouds differing by arbitrary scale factors
- Monotone-invariant: Robust to monotone transformations of dissimilarities
- Broadly applicable: Works with non-metric single-cell multiomics data
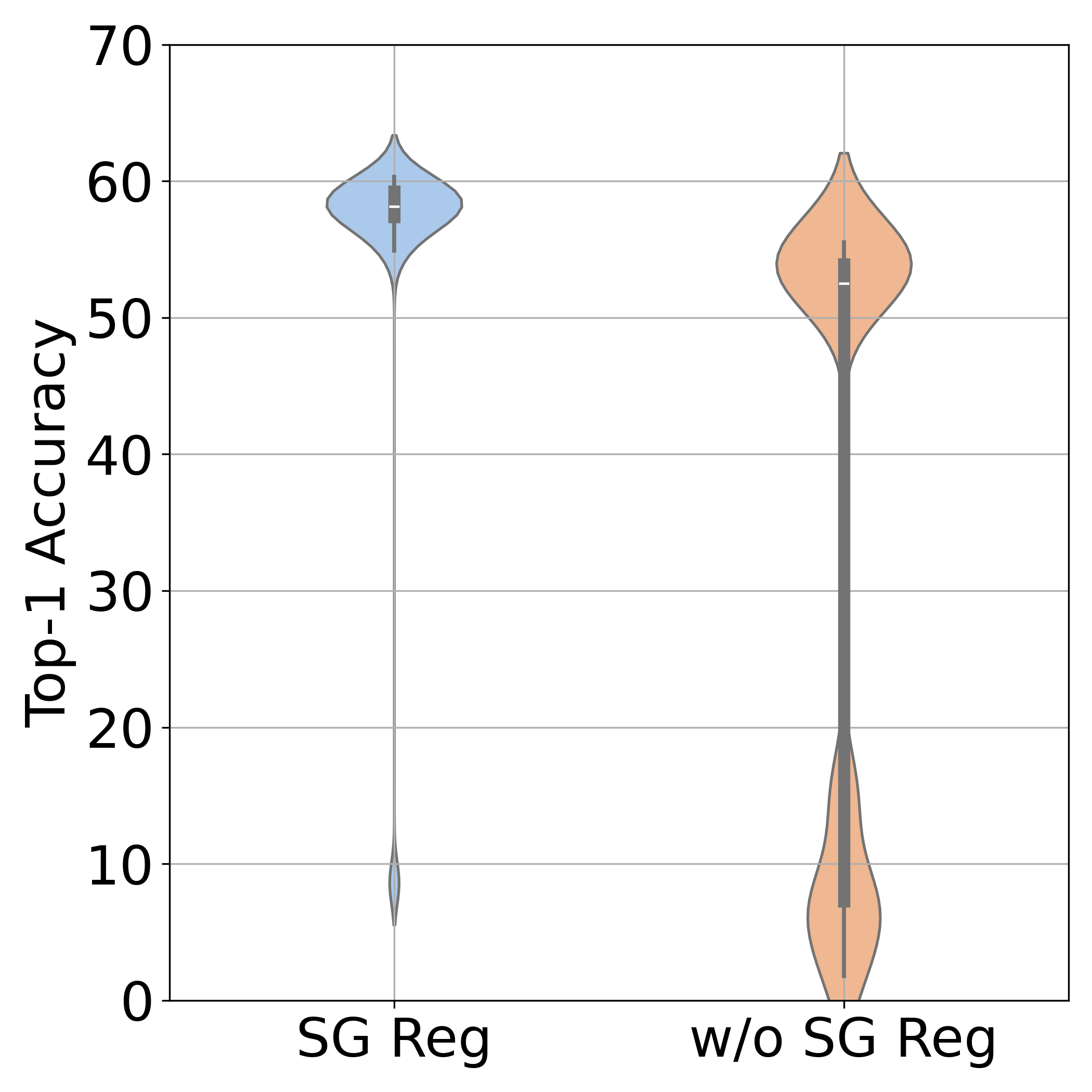
2. Spectral Geometric Regularization
We regularize the learned cost to be smooth on the product manifold:
\[\mathcal{E}_{\text{sm}} = \mathrm{trace}\left( \mathbf{C}^\T \mathbf{L}_{\mathcal{X}} \mathbf{C} + \mathbf{C} \mathbf{L}_\mathcal{Y} \mathbf{C}^\T \right)\]where $\mathbf{L}\mathcal{X}$ and $\mathbf{L}\mathcal{Y}$ are graph Laplacians. This encodes the intuition: similar samples in one domain should have similar costs relative to the other domain.
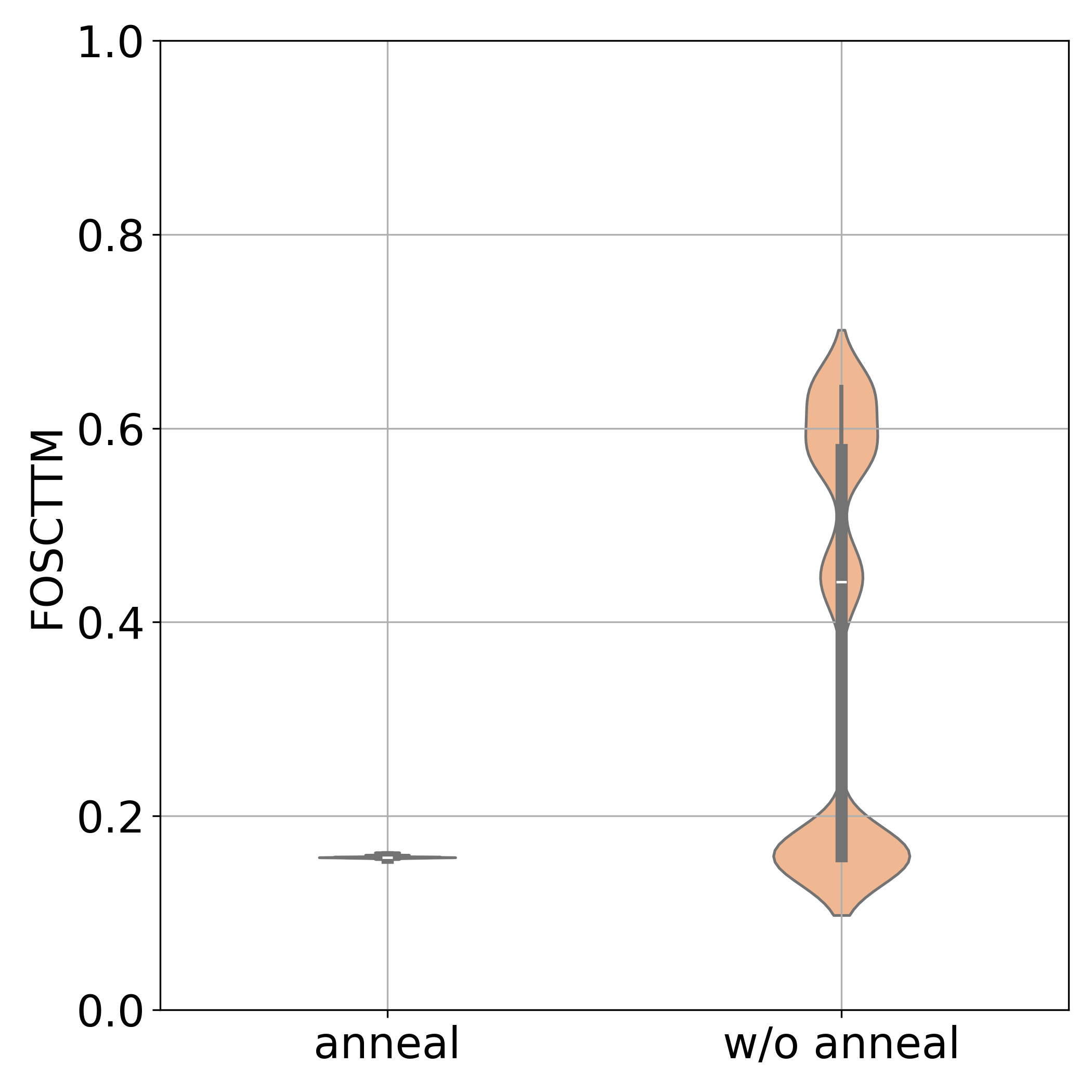
3. Simulated Annealing of Entropic Regularization
To avoid bad local minima, we gradually decrease the entropy regularization $\epsilon$ during training:
- High $\epsilon$ initially: Soft assignments provide global structure
- Low $\epsilon$ at end: Sharp assignments for precise alignment
- Coarse-to-fine refinement: Similar to multi-scale kernel matching in shape correspondence
Experiments
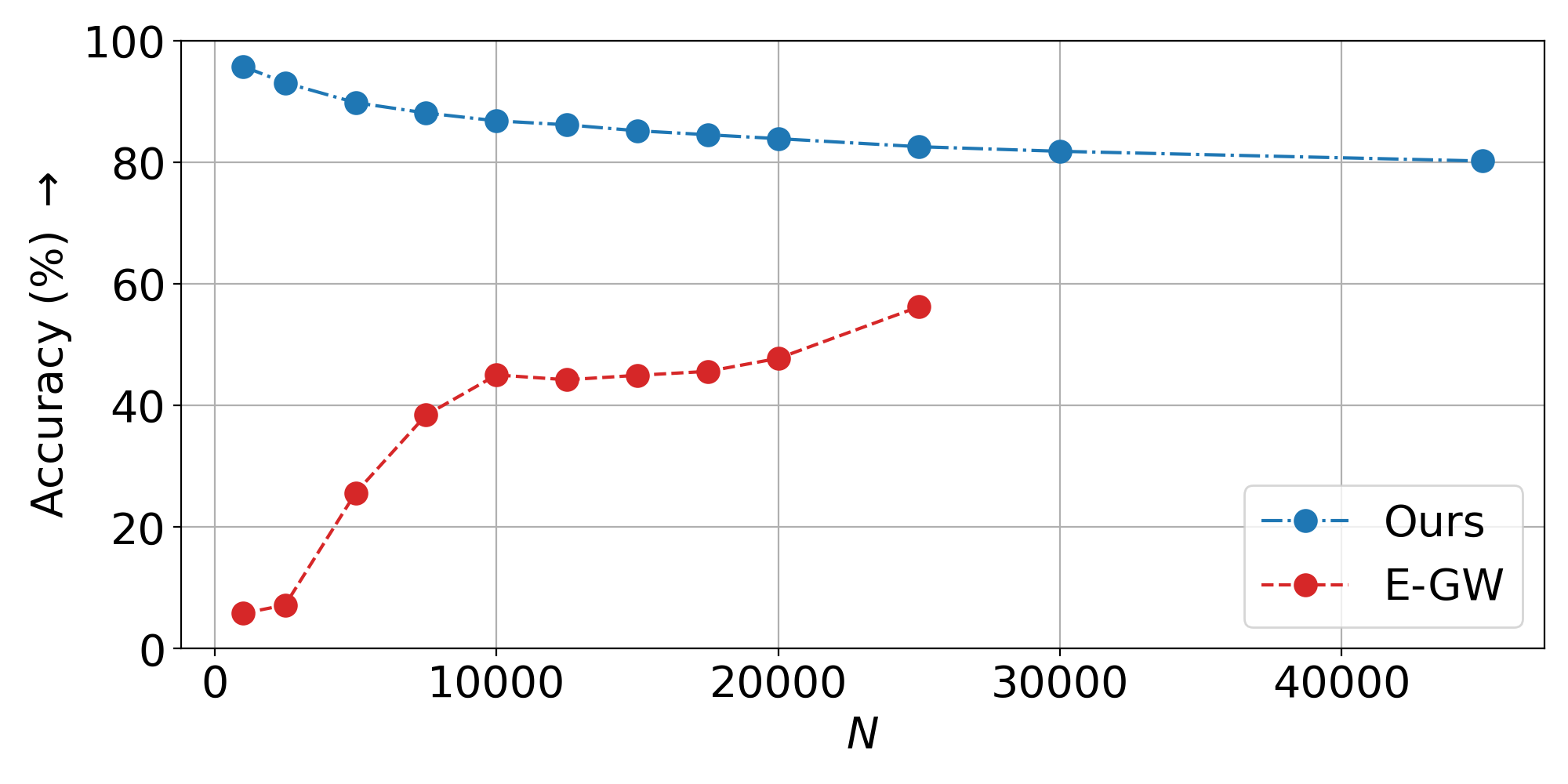
Scalability and Inductivity
We evaluate on CIFAR100 vision transformer embeddings under isometric (orthogonal) and non-isometric (rescaling) transformations.
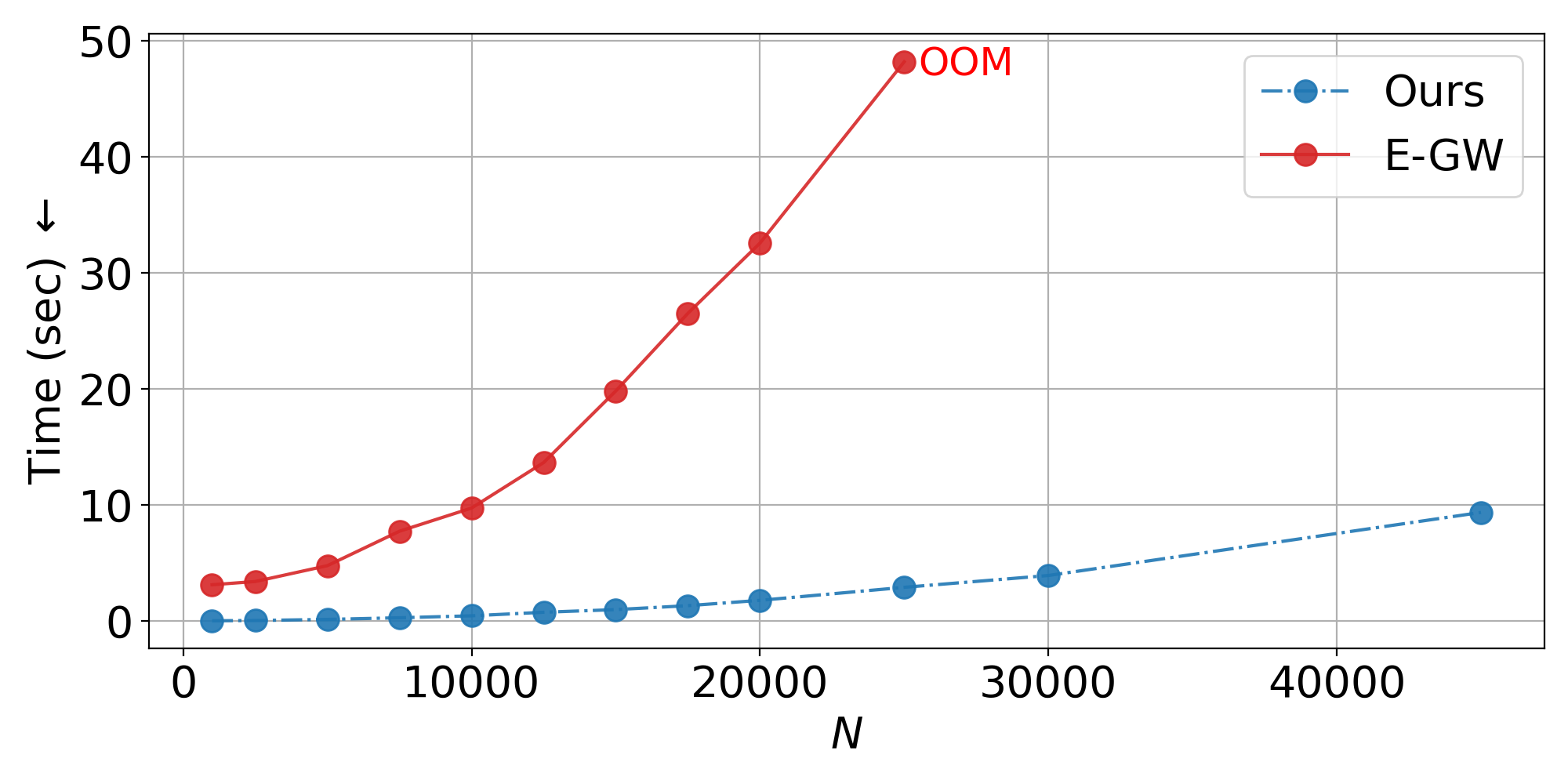
Isometric Setting: Training on 200 samples for 12 seconds, we evaluate inductively on up to 45,000 samples.
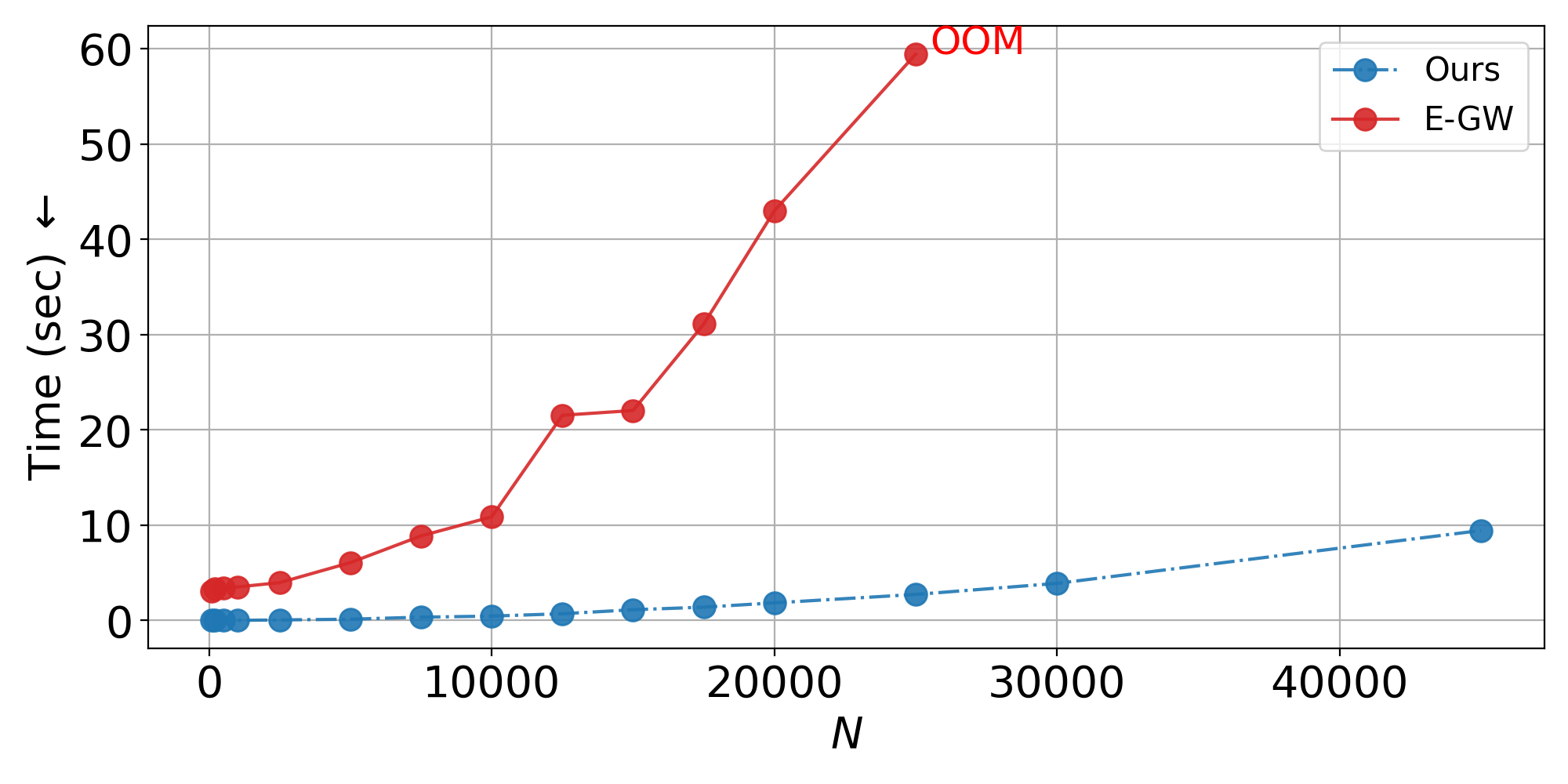
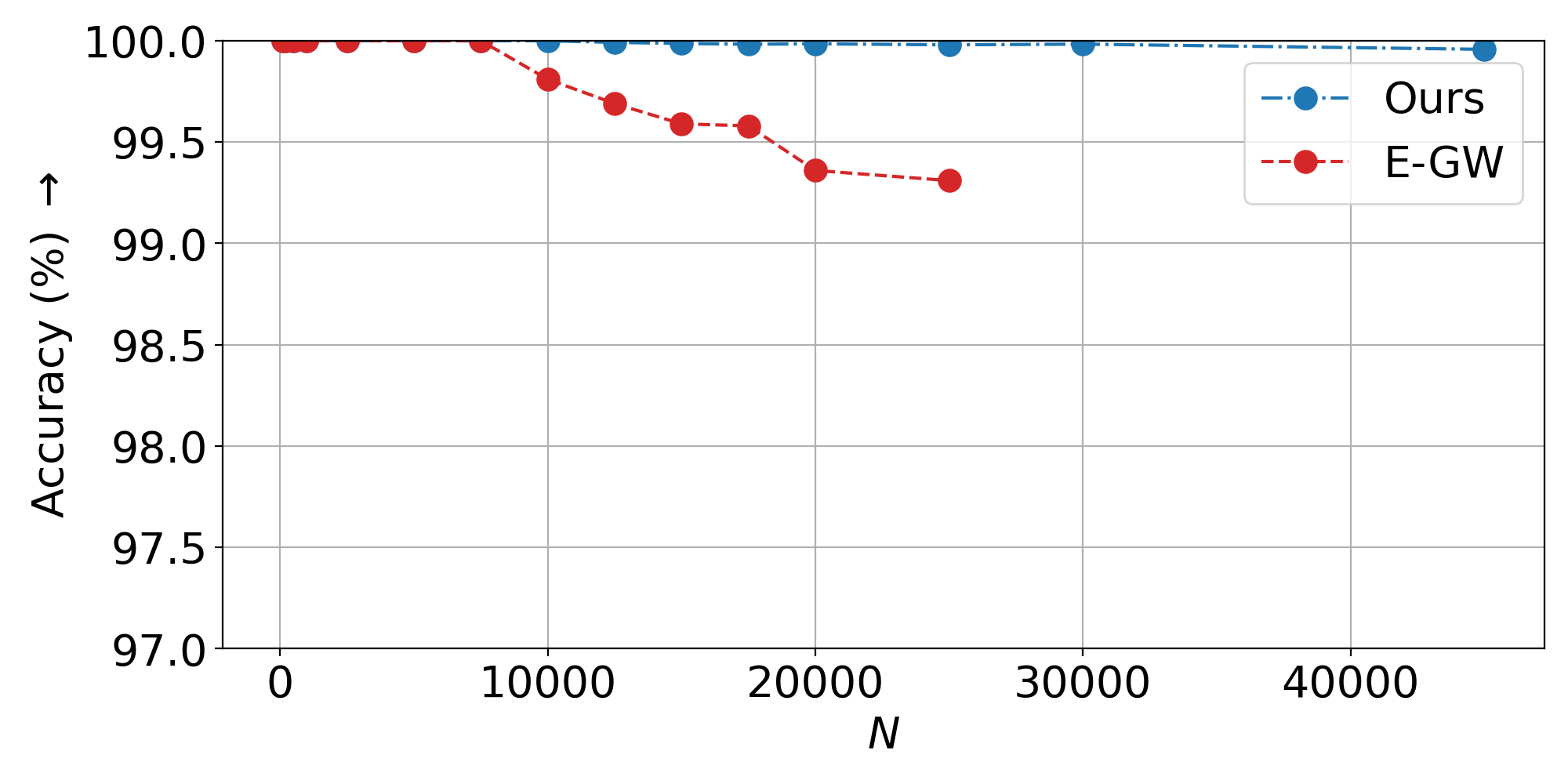
Non-Isometric Setting: ViT embeddings from rescaled images (384x384 vs 256x256), trained on 1,000 samples.
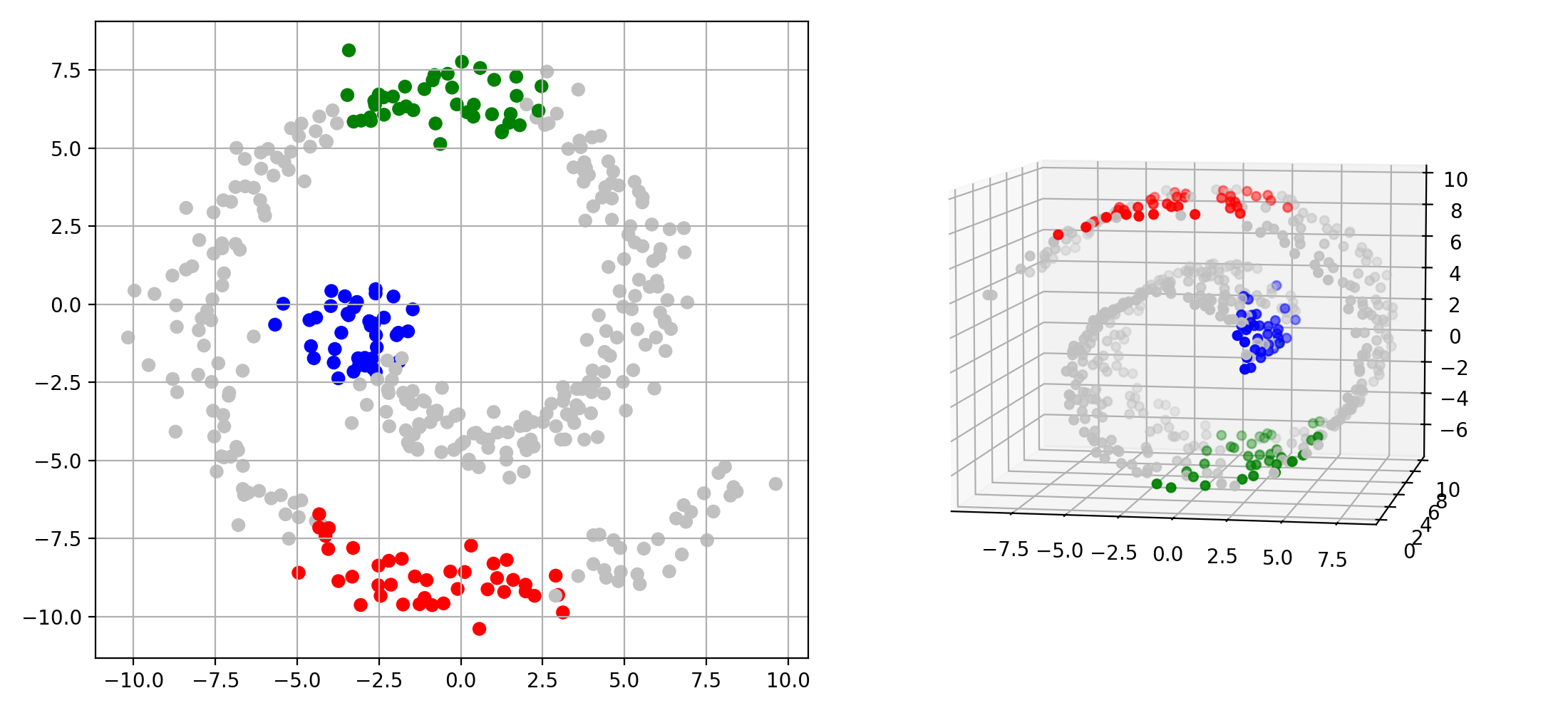
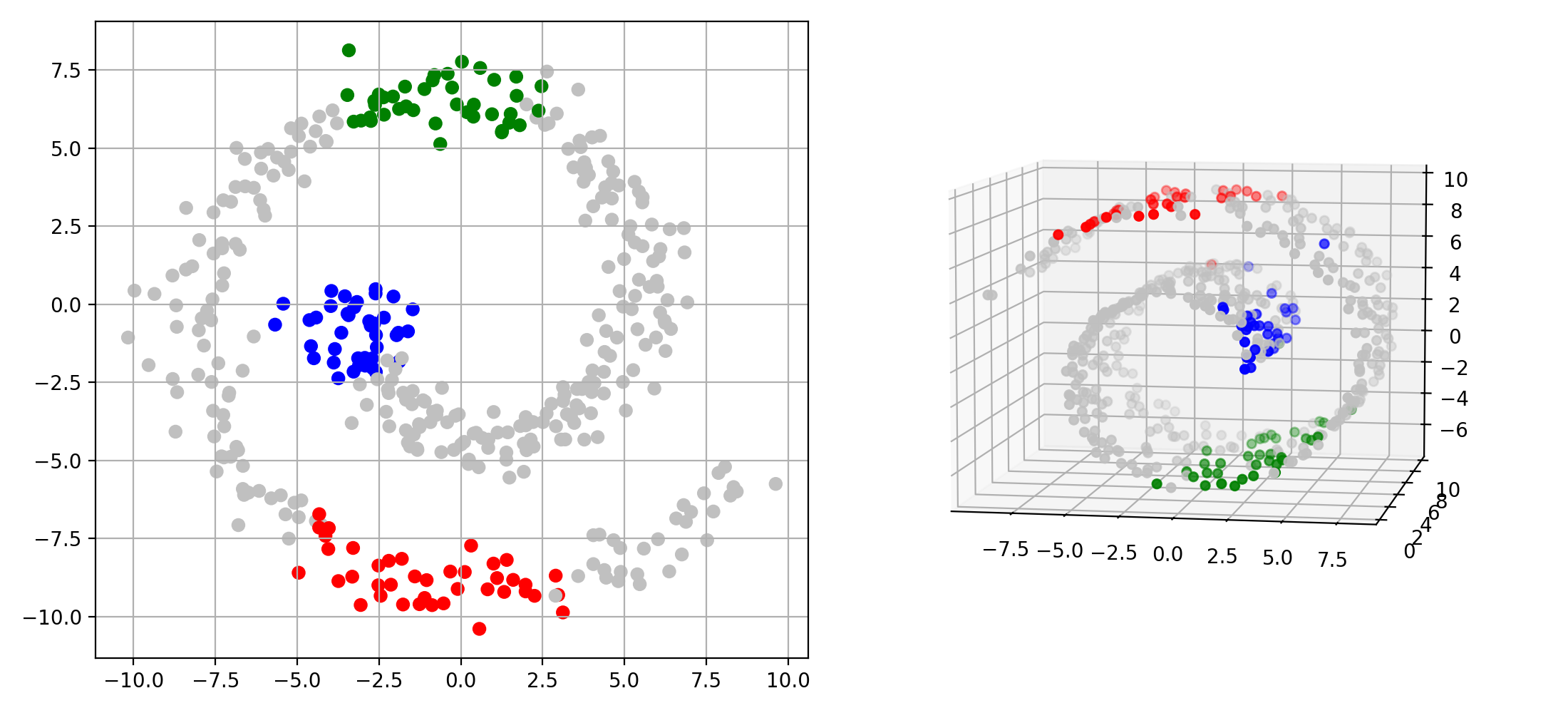
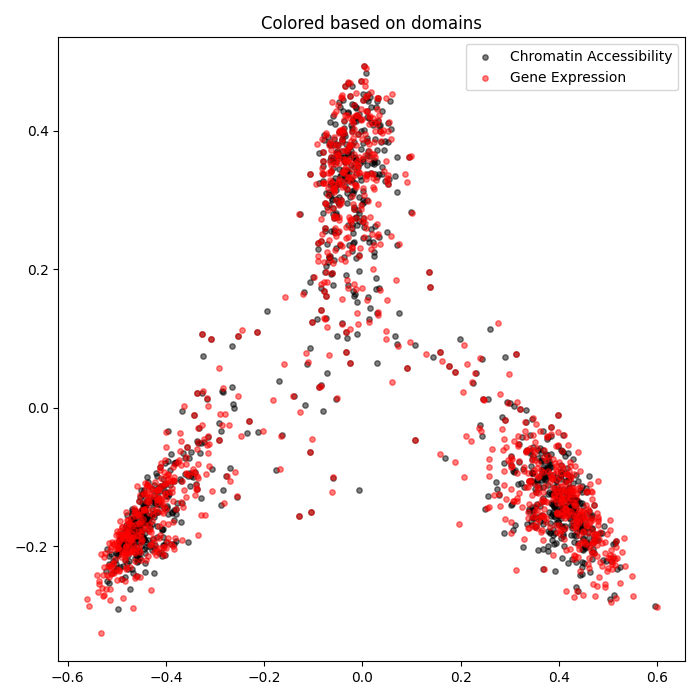
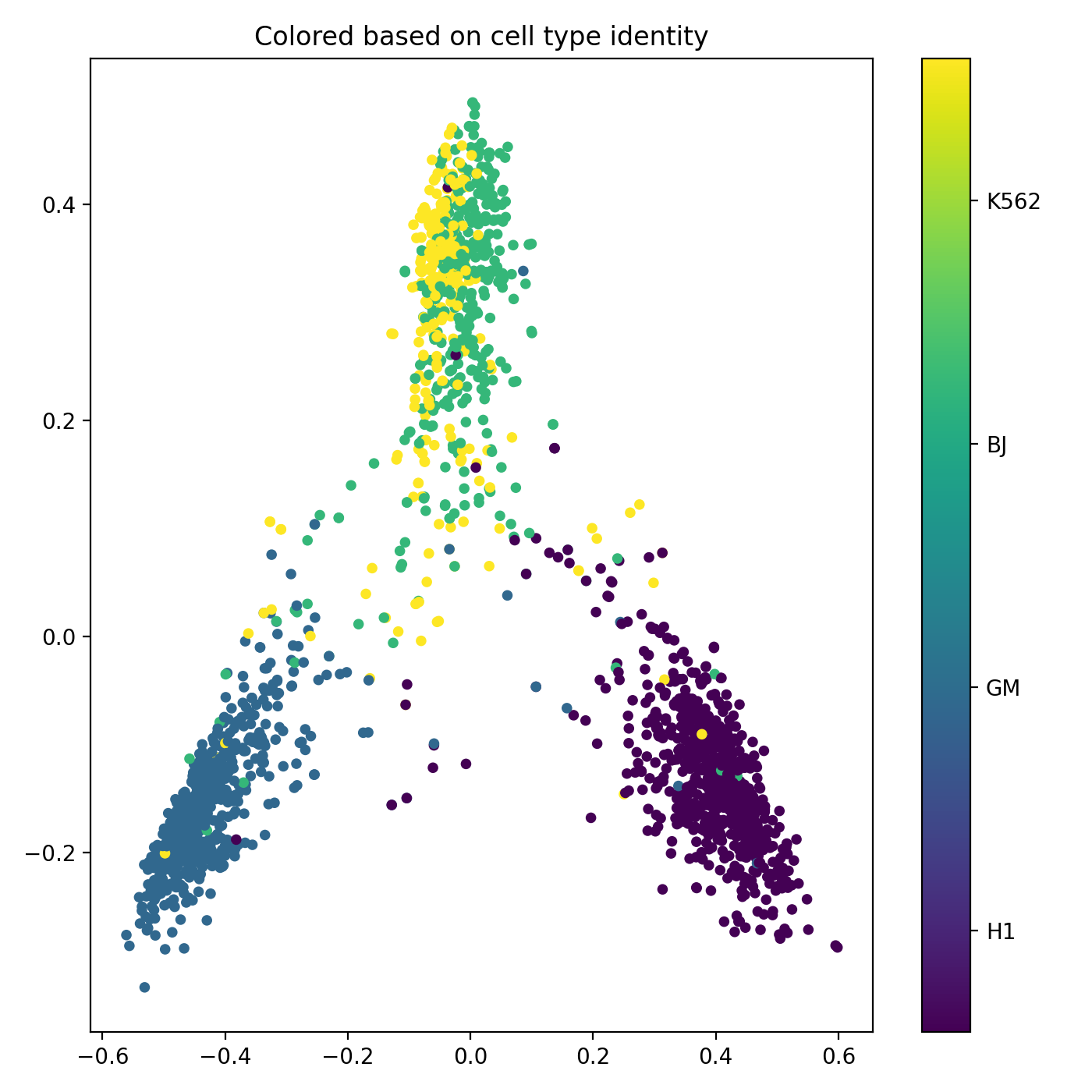
Single-Cell Multiomics Alignment
scSNARE-seq: Aligning RNA (gene expression) and ATAC (chromatin accessibility) from 1,047 cells across 4 cell lines.
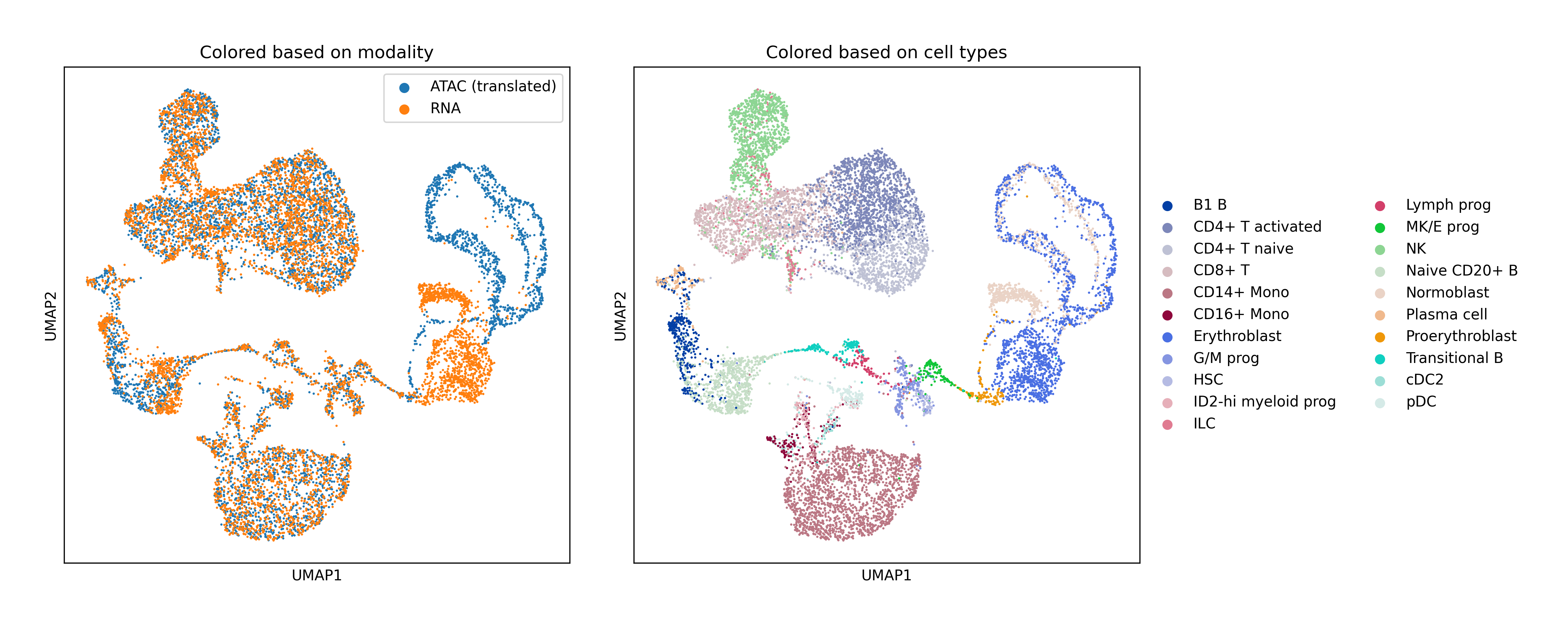
Human Bone Marrow: Large-scale alignment of RNA-seq and ATAC-seq with extensive hyperparameter search for fair baseline comparison.
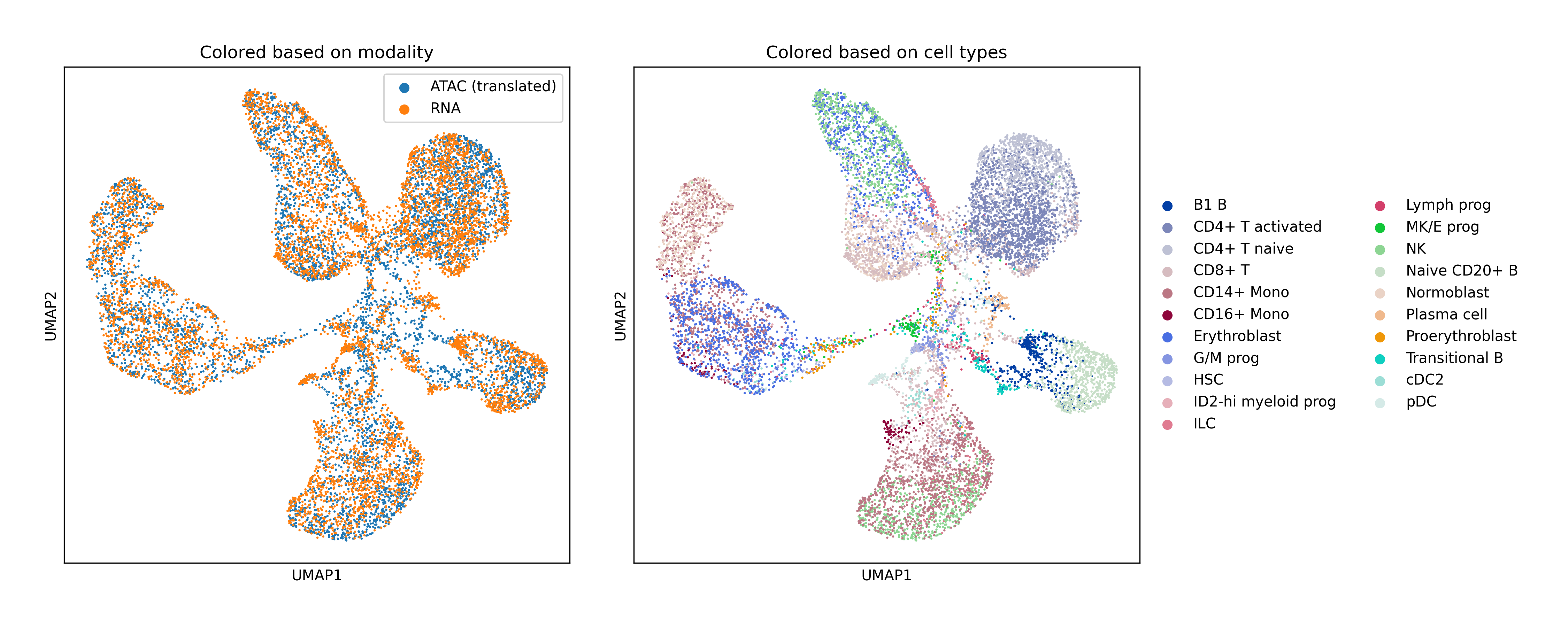
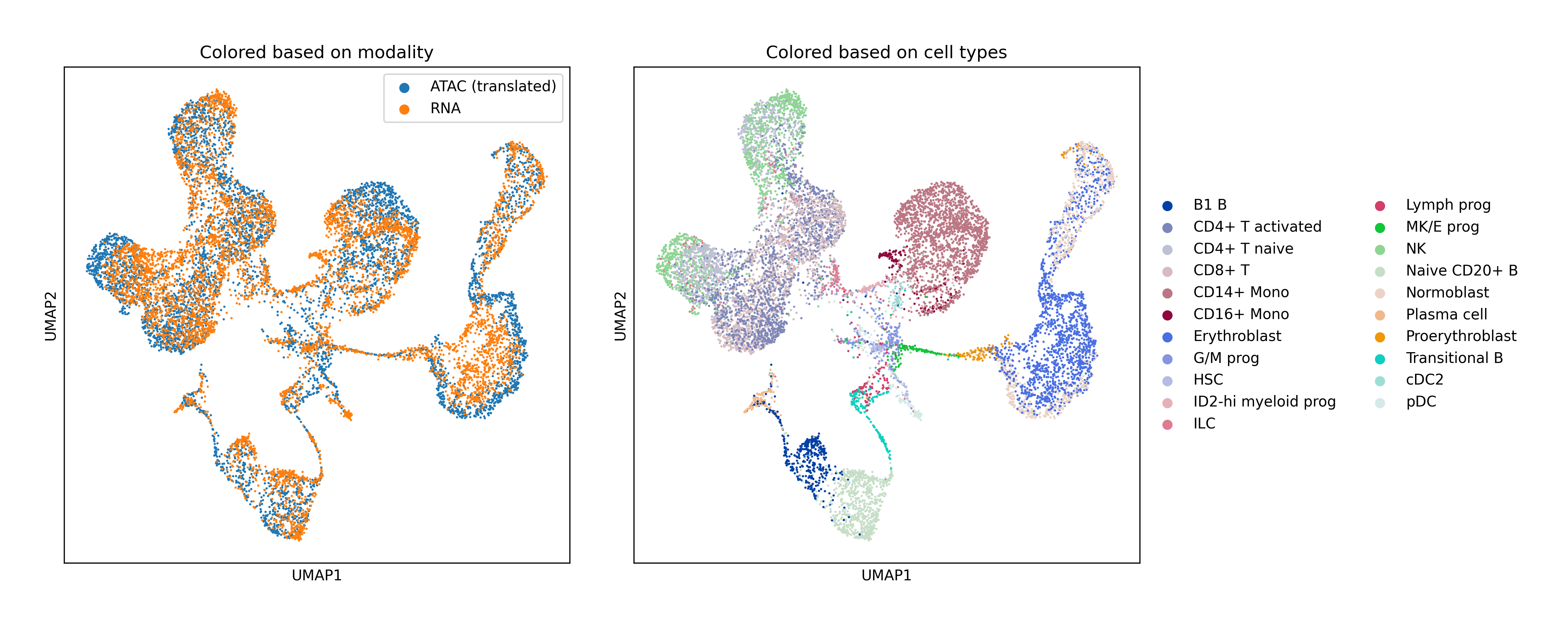
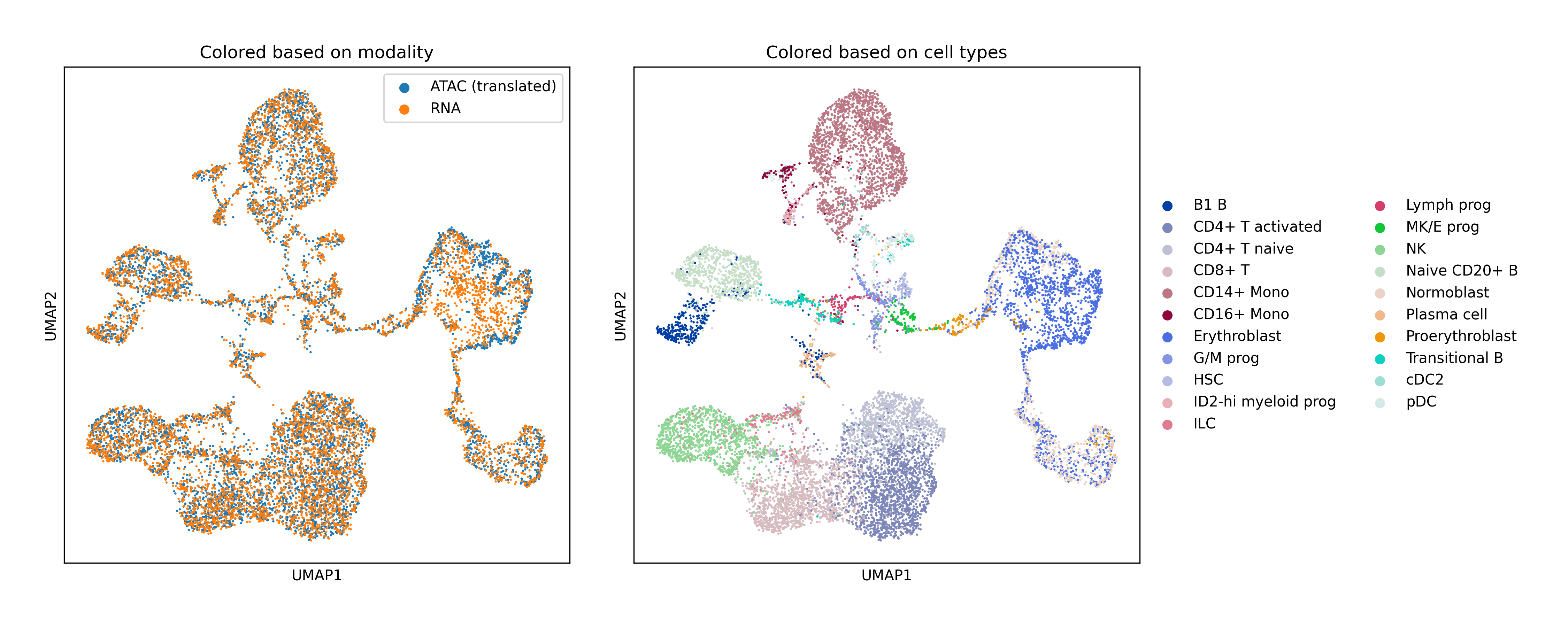
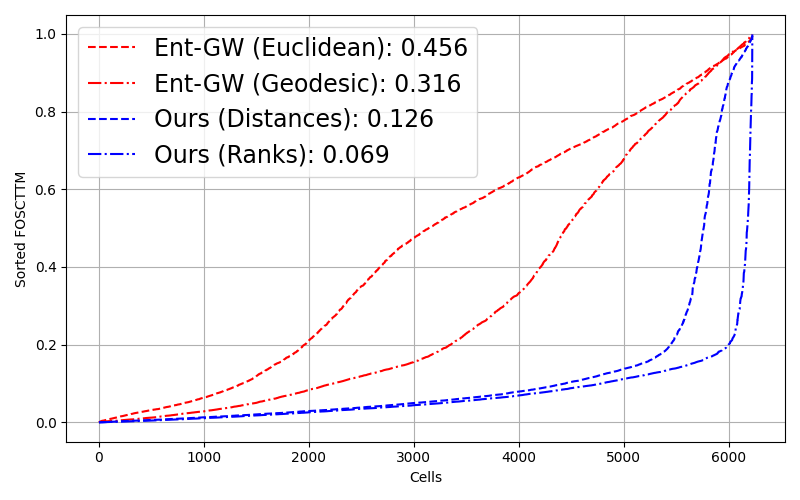
Key Findings
-
Inductive scalability: Train on hundreds of samples, generalize to tens of thousands—nearly 2x beyond the memory limit of standard entropic GW solvers.
-
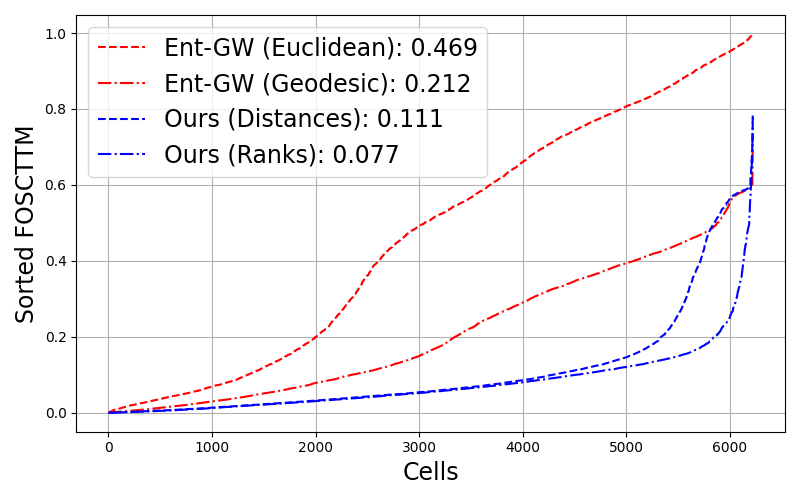
Rank-based matching wins: For real biological data with incompatible scales across modalities, matching distance ranks outperforms matching absolute distances. This extension enables alignment of truly non-metric structures.
-
Stabilization matters: Spectral regularization (+20% accuracy) and simulated annealing (near-zero variance) are crucial for reliable performance. These techniques break symmetries and avoid poor local minima.
-
Amortization pays off: By learning embeddings once, inference requires only a single efficient OT solve—no geodesic computation, no GW iterations. Training time is amortized across all future alignment tasks.
-
State-of-the-art on multiomics: Outperforms entropic GW and SCOT on single-cell RNA-ATAC alignment tasks, especially at larger scales. The rank-based formulation is particularly effective for disparate biological modalities.
Visual Summary
The method achieves superior alignment quality through three key innovations visible in the results:
- Scalability plots: Linear runtime scaling to 45K+ samples vs. memory failure of baselines at 25K
- Regularization effects: Dramatic variance reduction and accuracy gains from spectral smoothness and annealing
- Biological validation: Clean modality mixing with preserved cell type structure in UMAP visualizations
- Quantitative superiority: Significantly lower FOSCTTM scores compared to all baselines on bone marrow data
Citation
@inproceedings{vedula2024scalable,
title = {Scalable unsupervised alignment of general metric and non-metric structures},
author = {Vedula, Sanketh and Maiorca, Valentino and Basile, Lorenzo and
Locatello, Francesco and Bronstein, Alex},
booktitle = {ICML AI4Science Workshop},
year = {2024},
url = {https://arxiv.org/abs/2406.13507}
}
Authors
Sanketh Vedula¹’² · Valentino Maiorca²’³ · Lorenzo Basile²’⁴ · Francesco Locatello² · Alex Bronstein¹’²
¹Technion, Israel · ²Institute of Science and Technology Austria · ³Sapienza University of Rome, Italy · ⁴University of Trieste, Italy